AI-driven modelling of coronavirus spike protein celebrated with Gordon Bell Special Prize
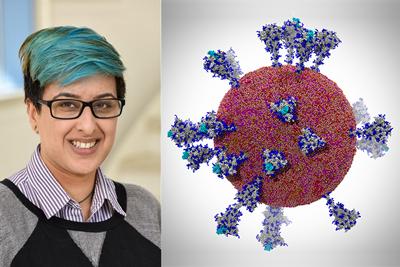
An international team of researchers have been recognised for successfully simulating the behaviour and vulnerabilities of a coronavirus in a first-of-its-kind feat in high performance computing.
The computational chemistry team, including the University of Southampton’s Professor Syma Khalid, have been awarded the Gordon Bell Special Prize for High Performance Computing-Based COVID-19 Research by the Association for Computing Machinery (ACM).
The study, led by UC San Diego’s Professor Rommie Amaro and Argonne National Laboratory’s (ANL) Dr Arvind Ramanathan, has investigated the movement and rearrangements in shape of the SARS-CoV-2 spike protein to understand how it triggers the process of infecting human cells.
The team were honoured at the virtual SC20, the International Conference for High Performance Computing, Networking, Storage, and Analysis this month.
Professor Khalid, Professor of Computational Biophysics at Southampton, says: “The project has predicted a key step in the process of the virus infecting our cells. The simulations were able to predict how the spike protein of the virus - the protein that latches on to our cells - will move and change shape, this is not possible to predict from static structures. If this change in shape can be prevented, then potentially infection can be prevented.”
Over 30 researchers contributed to the pioneering project as it built a workflow based on artificial intelligence to more efficiently simulate the spike.
Professor Khalid adds: “I am delighted to have played a small part in a large team effort lead by Rommie Amaro at UCSD. The team worked incredibly hard, and in particular the graduate students from UCSD showed a lot of dedication and maturity.
“It is gratifying to be able to contribute expertise I have been gained from my work on simulating bacterial membranes at Southampton, to help the team on their work on viral and eukaryotic membranes. We are still working together to simulate some of the other viral proteins, so we will hopefully have more to present soon.”
The results led to discoveries of one of the mechanisms that the virus uses to evade detection as well as a characterisation of interactions between the spike protein and a protein that the virus takes advantage of in human cells. This deeper understanding provided valuable insight the search for therapeutics or vaccines that might work to mitigate the virus.