Multiscale modelling and simulations to help design new drugs and vaccines to combat the coronavirus
Computational chemists at the University of Southampton are contributing to an international study that aims to facilitate the design of new antivirals and vaccines to combat SARS-COV2 (coronavirus).
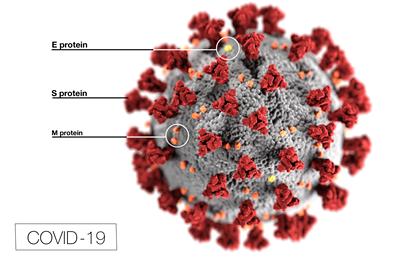
Professor Syma Khalid, from the School of Chemistry, is using multiscale modelling and simulations to understand and make predictions about a protein, known as the M protein, which is found within the membrane of the coronavirus.
The models will be used by a team at the University of California, San Diego, who are building an all-atom model of the entire virus.
It is hoped that the knowledge gained from the full model can help researchers design new drugs and vaccines to combat the coronavirus. The M protein is important as it is very abundant within the virus and protects the interior of the virus from the outside world. Therefore, in order for drugs to get inside the virus they must penetrate this layer of proteins.
Professor Khalid will be using coarse-grained molecular dynamics simulations to map the M protein movements. She explained: “The coronavirus is surrounded by a lipid membrane. The most abundant protein in the membrane is the M protein, however neither the structure of the protein nor the precise way it is arranged in the membrane, and how it interacts with other membrane proteins are known.”
The simulations are being performed on a combination of UK-leading high performance computing resources at Southampton and national facilities provided through UKRI.
“More information about this protein will help us understand how the virus survives, which will then help the search to find a treatment or vaccine. The models I create will be converted to all-atom resolution by Michael Feig at Michigan State University, and used in the larger model by Professor Rommie Amaro’s team in San Diego.”